Sensitivity analysis#
import os
from itertools import product
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import seaborn as sns
from h2ss import compare
cavern_diameter = np.arange(45, 106, step=5)
cavern_height = np.arange(80, 321, step=20)
def generate_sensitivity_data(diameter, height):
"""Generate data to perform sensitivity analysis"""
os.makedirs(os.path.join("data", "sensitivity"), exist_ok=True)
ds, extent, exclusions = compare.load_all_data()
for d, h in product(diameter, height):
filename = os.path.join(
"data", "sensitivity", f"sensitivity_d{d}_h{h}.csv"
)
if not os.path.isfile(filename):
df, _ = compare.capacity_function(ds, extent, exclusions, d, h)
df = df[["cavern_diameter", "cavern_height", "capacity"]]
df.to_csv(filename)
print(f"{filename} done!")
else:
print(f"{filename} exists!")
generate_sensitivity_data(cavern_diameter, cavern_height)
data/sensitivity/sensitivity_d45_h80.csv done!
data/sensitivity/sensitivity_d45_h100.csv done!
data/sensitivity/sensitivity_d45_h120.csv done!
data/sensitivity/sensitivity_d45_h140.csv done!
data/sensitivity/sensitivity_d45_h160.csv done!
data/sensitivity/sensitivity_d45_h180.csv done!
data/sensitivity/sensitivity_d45_h200.csv done!
data/sensitivity/sensitivity_d45_h220.csv done!
data/sensitivity/sensitivity_d45_h240.csv done!
data/sensitivity/sensitivity_d45_h260.csv done!
data/sensitivity/sensitivity_d45_h280.csv done!
data/sensitivity/sensitivity_d45_h300.csv done!
data/sensitivity/sensitivity_d45_h320.csv done!
data/sensitivity/sensitivity_d50_h80.csv done!
data/sensitivity/sensitivity_d50_h100.csv done!
data/sensitivity/sensitivity_d50_h120.csv done!
data/sensitivity/sensitivity_d50_h140.csv done!
data/sensitivity/sensitivity_d50_h160.csv done!
data/sensitivity/sensitivity_d50_h180.csv done!
data/sensitivity/sensitivity_d50_h200.csv done!
data/sensitivity/sensitivity_d50_h220.csv done!
data/sensitivity/sensitivity_d50_h240.csv done!
data/sensitivity/sensitivity_d50_h260.csv done!
data/sensitivity/sensitivity_d50_h280.csv done!
data/sensitivity/sensitivity_d50_h300.csv done!
data/sensitivity/sensitivity_d50_h320.csv done!
data/sensitivity/sensitivity_d55_h80.csv done!
data/sensitivity/sensitivity_d55_h100.csv done!
data/sensitivity/sensitivity_d55_h120.csv done!
data/sensitivity/sensitivity_d55_h140.csv done!
data/sensitivity/sensitivity_d55_h160.csv done!
data/sensitivity/sensitivity_d55_h180.csv done!
data/sensitivity/sensitivity_d55_h200.csv done!
data/sensitivity/sensitivity_d55_h220.csv done!
data/sensitivity/sensitivity_d55_h240.csv done!
data/sensitivity/sensitivity_d55_h260.csv done!
data/sensitivity/sensitivity_d55_h280.csv done!
data/sensitivity/sensitivity_d55_h300.csv done!
data/sensitivity/sensitivity_d55_h320.csv done!
data/sensitivity/sensitivity_d60_h80.csv done!
data/sensitivity/sensitivity_d60_h100.csv done!
data/sensitivity/sensitivity_d60_h120.csv done!
data/sensitivity/sensitivity_d60_h140.csv done!
data/sensitivity/sensitivity_d60_h160.csv done!
data/sensitivity/sensitivity_d60_h180.csv done!
data/sensitivity/sensitivity_d60_h200.csv done!
data/sensitivity/sensitivity_d60_h220.csv done!
data/sensitivity/sensitivity_d60_h240.csv done!
data/sensitivity/sensitivity_d60_h260.csv done!
data/sensitivity/sensitivity_d60_h280.csv done!
data/sensitivity/sensitivity_d60_h300.csv done!
data/sensitivity/sensitivity_d60_h320.csv done!
data/sensitivity/sensitivity_d65_h80.csv done!
data/sensitivity/sensitivity_d65_h100.csv done!
data/sensitivity/sensitivity_d65_h120.csv done!
data/sensitivity/sensitivity_d65_h140.csv done!
data/sensitivity/sensitivity_d65_h160.csv done!
data/sensitivity/sensitivity_d65_h180.csv done!
data/sensitivity/sensitivity_d65_h200.csv done!
data/sensitivity/sensitivity_d65_h220.csv done!
data/sensitivity/sensitivity_d65_h240.csv done!
data/sensitivity/sensitivity_d65_h260.csv done!
data/sensitivity/sensitivity_d65_h280.csv done!
data/sensitivity/sensitivity_d65_h300.csv done!
data/sensitivity/sensitivity_d65_h320.csv done!
data/sensitivity/sensitivity_d70_h80.csv done!
data/sensitivity/sensitivity_d70_h100.csv done!
data/sensitivity/sensitivity_d70_h120.csv done!
data/sensitivity/sensitivity_d70_h140.csv done!
data/sensitivity/sensitivity_d70_h160.csv done!
data/sensitivity/sensitivity_d70_h180.csv done!
data/sensitivity/sensitivity_d70_h200.csv done!
data/sensitivity/sensitivity_d70_h220.csv done!
data/sensitivity/sensitivity_d70_h240.csv done!
data/sensitivity/sensitivity_d70_h260.csv done!
data/sensitivity/sensitivity_d70_h280.csv done!
data/sensitivity/sensitivity_d70_h300.csv done!
data/sensitivity/sensitivity_d70_h320.csv done!
data/sensitivity/sensitivity_d75_h80.csv done!
data/sensitivity/sensitivity_d75_h100.csv done!
data/sensitivity/sensitivity_d75_h120.csv done!
data/sensitivity/sensitivity_d75_h140.csv done!
data/sensitivity/sensitivity_d75_h160.csv done!
data/sensitivity/sensitivity_d75_h180.csv done!
data/sensitivity/sensitivity_d75_h200.csv done!
data/sensitivity/sensitivity_d75_h220.csv done!
data/sensitivity/sensitivity_d75_h240.csv done!
data/sensitivity/sensitivity_d75_h260.csv done!
data/sensitivity/sensitivity_d75_h280.csv done!
data/sensitivity/sensitivity_d75_h300.csv done!
data/sensitivity/sensitivity_d75_h320.csv done!
data/sensitivity/sensitivity_d80_h80.csv done!
data/sensitivity/sensitivity_d80_h100.csv done!
data/sensitivity/sensitivity_d80_h120.csv done!
data/sensitivity/sensitivity_d80_h140.csv done!
data/sensitivity/sensitivity_d80_h160.csv done!
data/sensitivity/sensitivity_d80_h180.csv done!
data/sensitivity/sensitivity_d80_h200.csv done!
data/sensitivity/sensitivity_d80_h220.csv done!
data/sensitivity/sensitivity_d80_h240.csv done!
data/sensitivity/sensitivity_d80_h260.csv done!
data/sensitivity/sensitivity_d80_h280.csv done!
data/sensitivity/sensitivity_d80_h300.csv done!
data/sensitivity/sensitivity_d80_h320.csv done!
data/sensitivity/sensitivity_d85_h80.csv done!
data/sensitivity/sensitivity_d85_h100.csv done!
data/sensitivity/sensitivity_d85_h120.csv done!
data/sensitivity/sensitivity_d85_h140.csv done!
data/sensitivity/sensitivity_d85_h160.csv done!
data/sensitivity/sensitivity_d85_h180.csv done!
data/sensitivity/sensitivity_d85_h200.csv done!
data/sensitivity/sensitivity_d85_h220.csv done!
data/sensitivity/sensitivity_d85_h240.csv done!
data/sensitivity/sensitivity_d85_h260.csv done!
data/sensitivity/sensitivity_d85_h280.csv done!
data/sensitivity/sensitivity_d85_h300.csv done!
data/sensitivity/sensitivity_d85_h320.csv done!
data/sensitivity/sensitivity_d90_h80.csv done!
data/sensitivity/sensitivity_d90_h100.csv done!
data/sensitivity/sensitivity_d90_h120.csv done!
data/sensitivity/sensitivity_d90_h140.csv done!
data/sensitivity/sensitivity_d90_h160.csv done!
data/sensitivity/sensitivity_d90_h180.csv done!
data/sensitivity/sensitivity_d90_h200.csv done!
data/sensitivity/sensitivity_d90_h220.csv done!
data/sensitivity/sensitivity_d90_h240.csv done!
data/sensitivity/sensitivity_d90_h260.csv done!
data/sensitivity/sensitivity_d90_h280.csv done!
data/sensitivity/sensitivity_d90_h300.csv done!
data/sensitivity/sensitivity_d90_h320.csv done!
data/sensitivity/sensitivity_d95_h80.csv done!
data/sensitivity/sensitivity_d95_h100.csv done!
data/sensitivity/sensitivity_d95_h120.csv done!
data/sensitivity/sensitivity_d95_h140.csv done!
data/sensitivity/sensitivity_d95_h160.csv done!
data/sensitivity/sensitivity_d95_h180.csv done!
data/sensitivity/sensitivity_d95_h200.csv done!
data/sensitivity/sensitivity_d95_h220.csv done!
data/sensitivity/sensitivity_d95_h240.csv done!
data/sensitivity/sensitivity_d95_h260.csv done!
data/sensitivity/sensitivity_d95_h280.csv done!
data/sensitivity/sensitivity_d95_h300.csv done!
data/sensitivity/sensitivity_d95_h320.csv done!
data/sensitivity/sensitivity_d100_h80.csv done!
data/sensitivity/sensitivity_d100_h100.csv done!
data/sensitivity/sensitivity_d100_h120.csv done!
data/sensitivity/sensitivity_d100_h140.csv done!
data/sensitivity/sensitivity_d100_h160.csv done!
data/sensitivity/sensitivity_d100_h180.csv done!
data/sensitivity/sensitivity_d100_h200.csv done!
data/sensitivity/sensitivity_d100_h220.csv done!
data/sensitivity/sensitivity_d100_h240.csv done!
data/sensitivity/sensitivity_d100_h260.csv done!
data/sensitivity/sensitivity_d100_h280.csv done!
data/sensitivity/sensitivity_d100_h300.csv done!
data/sensitivity/sensitivity_d100_h320.csv done!
data/sensitivity/sensitivity_d105_h80.csv done!
data/sensitivity/sensitivity_d105_h100.csv done!
data/sensitivity/sensitivity_d105_h120.csv done!
data/sensitivity/sensitivity_d105_h140.csv done!
data/sensitivity/sensitivity_d105_h160.csv done!
data/sensitivity/sensitivity_d105_h180.csv done!
data/sensitivity/sensitivity_d105_h200.csv done!
data/sensitivity/sensitivity_d105_h220.csv done!
data/sensitivity/sensitivity_d105_h240.csv done!
data/sensitivity/sensitivity_d105_h260.csv done!
data/sensitivity/sensitivity_d105_h280.csv done!
data/sensitivity/sensitivity_d105_h300.csv done!
data/sensitivity/sensitivity_d105_h320.csv done!
filelist = []
for cd, ch in product(cavern_diameter, cavern_height):
filelist.append(
os.path.join("data", "sensitivity", f"sensitivity_d{cd}_h{ch}.csv")
)
cdf = pd.concat((pd.read_csv(f) for f in filelist), ignore_index=True)
cdf.drop(columns=["Unnamed: 0"], inplace=True)
cdf["cavern_height"] = cdf["cavern_height"].astype(int)
cdf.describe()
| cavern_diameter | cavern_height | capacity | |
|---|---|---|---|
| count | 31530.000000 | 31530.000000 | 31530.000000 |
| mean | 64.269109 | 125.683476 | 71.881045 |
| std | 17.312295 | 51.186041 | 54.891919 |
| min | 45.000000 | 80.000000 | 10.663307 |
| 25% | 50.000000 | 80.000000 | 35.064305 |
| 50% | 60.000000 | 100.000000 | 55.737384 |
| 75% | 75.000000 | 140.000000 | 89.742942 |
| max | 105.000000 | 320.000000 | 511.809957 |
# cavern height-to-diameter-ratio
pd.Series((cdf["cavern_height"] / cdf["cavern_diameter"]).unique()).describe()
count 130.000000
mean 2.908468
std 1.437914
min 0.761905
25% 1.783333
50% 2.701754
75% 3.685897
max 7.111111
dtype: float64
ax = sns.scatterplot(
data=cdf,
hue="cavern_diameter",
y="capacity",
x="cavern_height",
linewidth=0,
alpha=0.75,
s=5,
)
sns.despine()
plt.show()
len(cdf["cavern_diameter"].unique()) == len(cdf["cavern_height"].unique())
True
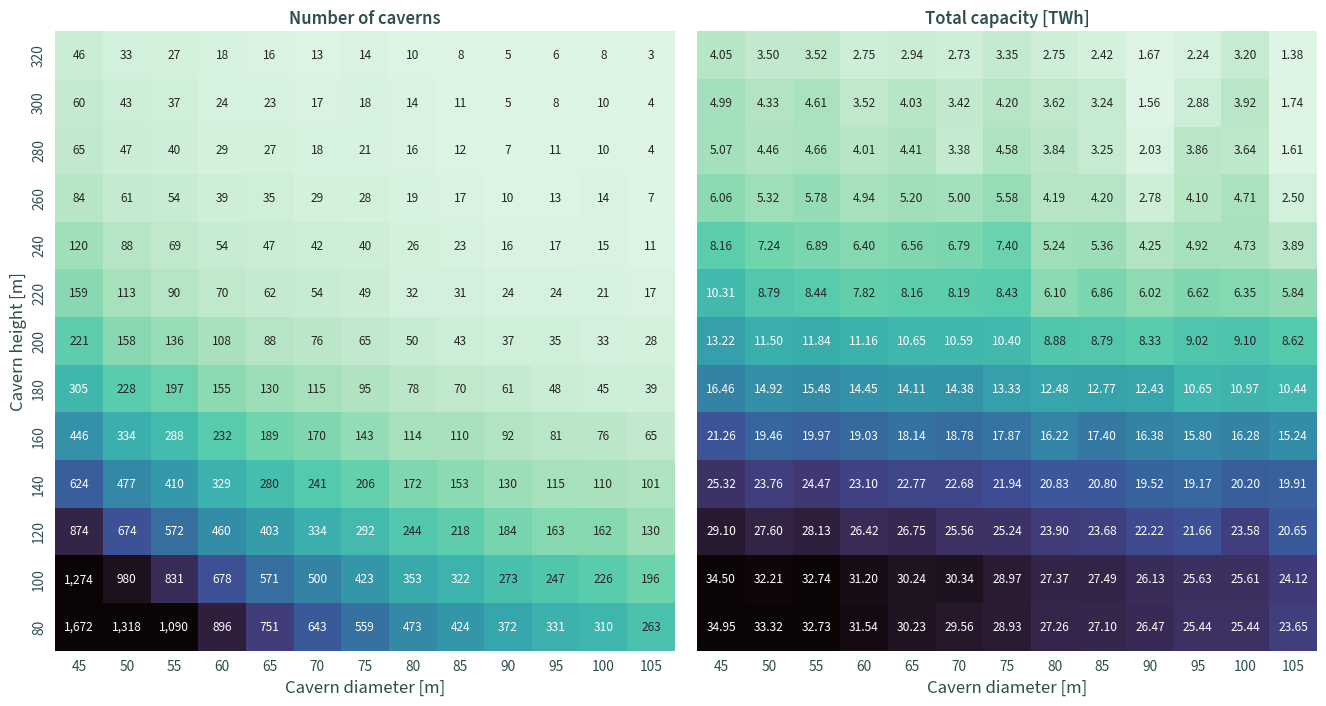
Number of caverns and total capacity#
f, ax = plt.subplots(1, 2, figsize=(14, 7), sharey=True)
data = (
cdf.groupby(["cavern_height", "cavern_diameter"])
.count()
.reset_index()
.pivot(index="cavern_height", columns="cavern_diameter", values="capacity")
.sort_index(ascending=False)
)
sns.heatmap(
data,
ax=ax[0],
cmap="mako_r",
cbar=False,
square=True,
annot=True,
fmt=",d",
robust=True,
)
data = cdf.copy()
data["capacity"] = data["capacity"] / 1000
data = (
data.groupby(["cavern_height", "cavern_diameter"])
.sum()
.reset_index()
.pivot(index="cavern_height", columns="cavern_diameter", values="capacity")
.sort_index(ascending=False)
)
sns.heatmap(
data,
ax=ax[1],
cmap="mako_r",
cbar=False,
square=True,
annot=True,
robust=True,
fmt=".2f",
)
for a in ax.flat:
a.tick_params(axis="y", labelsize=11.5, left=False)
a.tick_params(axis="x", labelsize=11.5, bottom=False)
a.set_xlabel("Cavern diameter [m]")
ax[0].set_ylabel("Cavern height [m]")
ax[1].set_ylabel("")
ax[0].set_title("Number of caverns")
ax[1].set_title("Total capacity [TWh]")
plt.tight_layout()
# plt.savefig(
# os.path.join("graphics", "fig_sensitivity_heatmap.jpg"),
# format="jpg",
# dpi=600,
# )
plt.show()
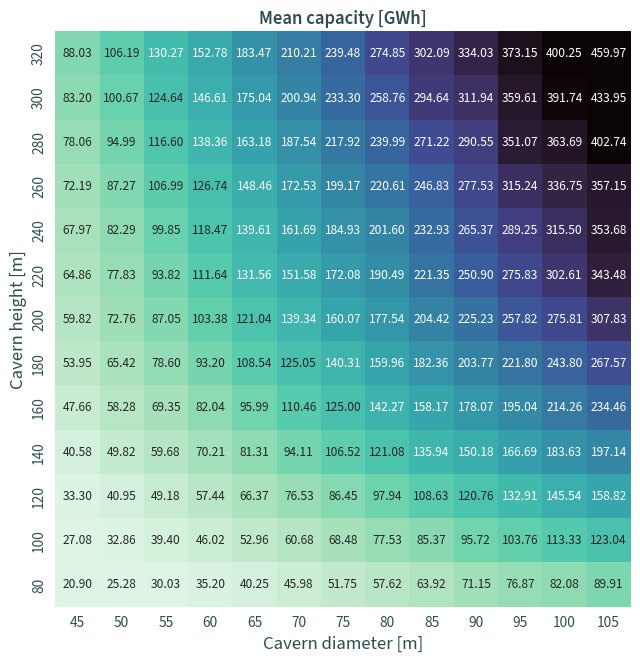
Mean capacity#
data = (
cdf.groupby(["cavern_height", "cavern_diameter"])
.mean()
.reset_index()
.pivot(index="cavern_height", columns="cavern_diameter", values="capacity")
.sort_index(ascending=False)
)
f, ax = plt.subplots(figsize=(7, 7))
sns.heatmap(
data,
ax=ax,
cmap="mako_r",
# cbar_kws={"label": "Mean capacity [GWh]"},
cbar=False,
square=True,
annot=True,
robust=True,
fmt=".2f",
)
ax.set_xlabel("Cavern diameter [m]")
ax.set_ylabel("Cavern height [m]")
ax.tick_params(axis="y", labelsize=11.5, left=False)
ax.tick_params(axis="x", labelsize=11.5, bottom=False)
plt.title("Mean capacity [GWh]")
# ax.plot(8.5, 10.5, "*", markersize=10, color="black")
plt.tight_layout()
plt.show()
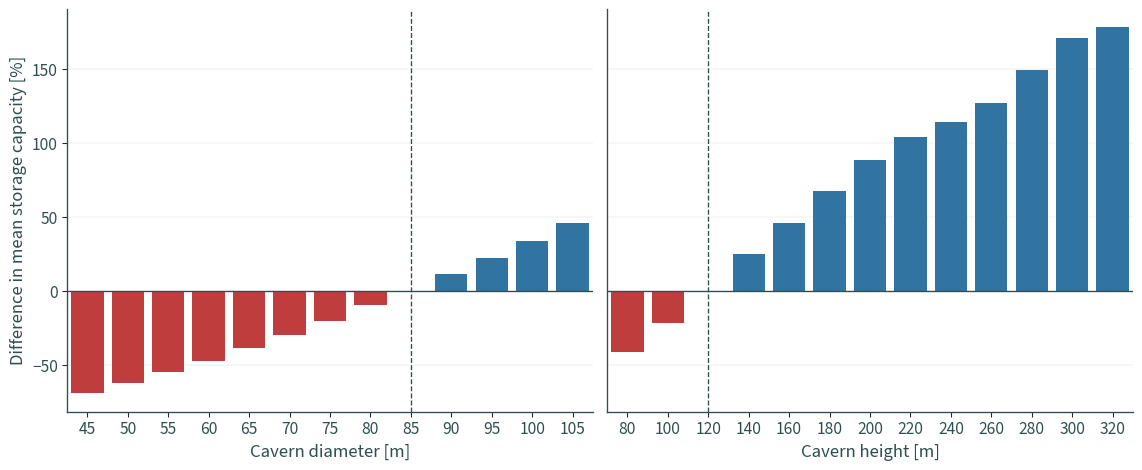
Base case#
base = cdf[
(cdf["cavern_diameter"] == 85) & (cdf["cavern_height"] == 120)
].reset_index(drop=True)
base.describe()[["capacity"]]
| capacity | |
|---|---|
| count | 218.000000 |
| mean | 108.634041 |
| std | 25.946851 |
| min | 55.946359 |
| 25% | 89.482842 |
| 50% | 109.211632 |
| 75% | 129.493525 |
| max | 158.798899 |
base_mean = base[["capacity"]].mean().values[0]
base_sum = base[["capacity"]].sum().values[0]
base_count = base[["capacity"]].count().values[0]
print(f"{base_mean:.3f}")
108.634
print(f"{base_sum:.3f}")
23682.221
base_count
218
Base diameter, varying height#
dd = cdf[(cdf["cavern_diameter"] == 85)].reset_index(drop=True)
dd_mean = (
pd.DataFrame(dd.groupby("cavern_height").mean()["capacity"] - base_mean)
/ base_mean
* 100
).reset_index()
dd_sum = (
pd.DataFrame(dd.groupby("cavern_height").sum()["capacity"] - base_sum)
/ base_sum
* 100
).reset_index()
dd_count = (
pd.DataFrame(dd.groupby("cavern_height").count()["capacity"] - base_count)
/ base_count
* 100
).reset_index()
Base height, varying diameter#
dh = cdf[(cdf["cavern_height"] == 120)].reset_index(drop=True)
dh_mean = (
pd.DataFrame(dh.groupby("cavern_diameter").mean()["capacity"] - base_mean)
/ base_mean
* 100
).reset_index()
dh_sum = (
pd.DataFrame(dh.groupby("cavern_diameter").sum()["capacity"] - base_sum)
/ base_sum
* 100
).reset_index()
dh_count = (
pd.DataFrame(
dh.groupby("cavern_diameter").count()["capacity"] - base_count
)
/ base_count
* 100
).reset_index()
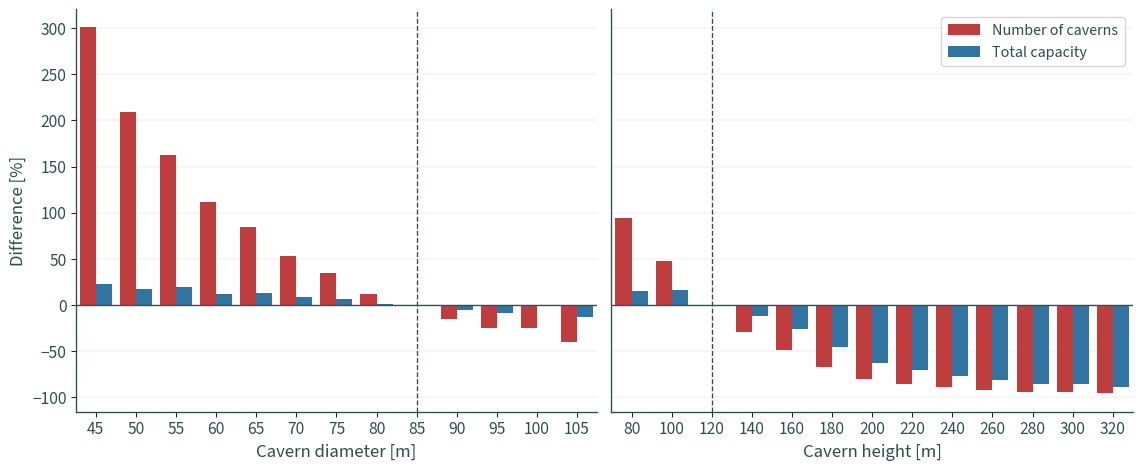
Combined plots#
dh_sum["type"] = "Total capacity"
dh_count["type"] = "Number of caverns"
dd_sum["type"] = "Total capacity"
dd_count["type"] = "Number of caverns"
f, ax = plt.subplots(1, 2, figsize=(12, 5), sharey=True)
sns.barplot(
data=pd.concat([dh_count, dh_sum]),
hue="type",
y="capacity",
x="cavern_diameter",
palette=sns.color_palette(["tab:red", "tab:blue"]),
legend=False,
ax=ax[0],
)
sns.barplot(
data=pd.concat([dd_count, dd_sum]),
hue="type",
y="capacity",
x="cavern_height",
palette=sns.color_palette(["tab:red", "tab:blue"]),
ax=ax[1],
)
for a in ax.flat:
a.axhline(0, color="darkslategrey", linewidth=1)
a.yaxis.grid(True, linewidth=0.25)
# a.set(ylim=(-100, 200))
ax[0].axvline("85", color="darkslategrey", linewidth=1, linestyle="dashed")
ax[1].axvline("120", color="darkslategrey", linewidth=1, linestyle="dashed")
ax[0].set_xlabel("Cavern diameter [m]")
ax[1].set_xlabel("Cavern height [m]")
ax[0].set_ylabel("Difference [%]")
ax[1].legend(title=None, fontsize=12)
ax[1].tick_params(axis="y", left=False)
sns.despine()
plt.tight_layout()
# plt.savefig(
# os.path.join("graphics", "Figure_9.png"),
# format="png",
# dpi=600,
# )
plt.show()
def colour_label(dataset):
"Use different colours for negative and positive values"
conditions = [(dataset["capacity"] < 0), (dataset["capacity"] >= 0)]
choices = ["N", "P"]
dataset["colour"] = np.select(conditions, choices)
return dataset
f, ax = plt.subplots(1, 2, figsize=(12, 5), sharey=True)
sns.barplot(
data=colour_label(dh_mean),
y="capacity",
x="cavern_diameter",
hue="colour",
palette=sns.color_palette(["tab:red", "tab:blue"]),
legend=False,
ax=ax[0],
)
sns.barplot(
data=colour_label(dd_mean),
y="capacity",
x="cavern_height",
hue="colour",
palette=sns.color_palette(["tab:red", "tab:blue"]),
legend=False,
ax=ax[1],
)
for a in ax.flat:
a.axhline(0, color="darkslategrey", linewidth=1)
a.yaxis.grid(True, linewidth=0.25)
ax[0].axvline("85", color="darkslategrey", linewidth=1, linestyle="dashed")
ax[1].axvline("120", color="darkslategrey", linewidth=1, linestyle="dashed")
ax[0].set_xlabel("Cavern diameter [m]")
ax[1].set_xlabel("Cavern height [m]")
ax[0].set_ylabel("Difference in mean storage capacity [%]")
ax[1].tick_params(axis="y", left=False)
sns.despine()
plt.tight_layout()
plt.show()